
* Factor analytic model assumes that the data comes from a well-defined model where the underlying factors satisfy assumptions mentioned before.
* PCA transforms the variables to principal components. * Principal component analysis (PCA) is a transformation of the data. The two are the same under certain case in fact (can you tell when?). # How is this different principal component analysis? Constrain upper triangle of loading matrix to zero. * If *k=1*, then there is no constraint applied and no rotation is necessary.ġ. * A square matrix **Q** of size *t* is orthogonal if the second accounts for the next largest amount of estimated genetic covariance and so on. the first rotated factor accounts for the maximum amount of estimated genetic covariance, The approach we use in the practical session, we use an approach similar to principal components, in that: * In fact, the rotated loadings and factor are also solutions. * If we do not impose some constraint as per slide 5, then there are many possible solutions for factor loadings. * Suppose that we have an orthogonal matrix **Q** of size *t*. * The first approach is akin to using coefficient of determination *R2* in linear regression.

(2018) Order Selection and Sparsity in Latent Variable Models via the Ordered Factor LASSO. You can use OFAL penalty proposed in Hui et al. You can use a hypothesis testing approach or use of information criterion.ģ. Pragmatically, you can use some threshold for overall percentage of between genetic variances explained by the *k* factors:Ģ.
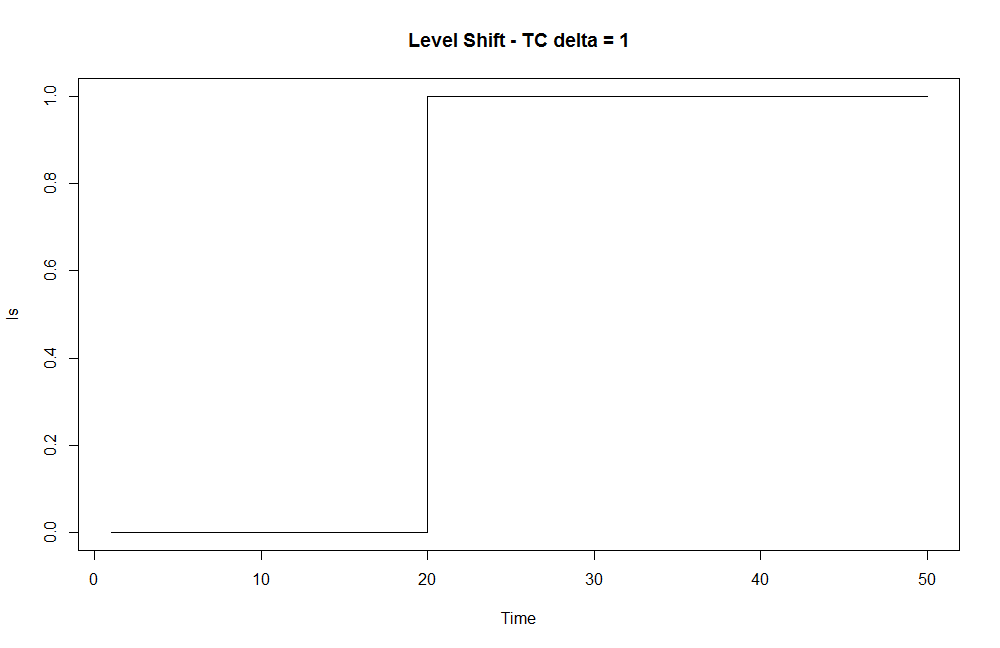
ASREML OUTLIERS HOW TO
# How to choose the order, *k*, of FA model?ġ. * The specific factor represent an effect specific to that environment. * The common factor represent how the genotype responds to that covariate. * The loadings represent some latent environmental covariate.
* Notice that this is like a linear regression model except the covariates are estimated from the data. * Note: our FA model is different to the standard FA model due to the separable structure of **G**ge. * FA Model is a special case of latent variable model when the responses are conditionally normally distributed. * *Trial*, *site* and *environment* are used synonymously. # Condition to use FA model over US model * As FA model is to offer a simpler model then it does not make sense to have more parameters to estimate in FA model than the US model. * The number of variance parameters to estimate for FA model grows. # The number of variance parameters to estimate * `asreml` constrains such that the upper triangle of the loading matrix are zeroes.
* Due to identifiability, some contraints are applied to the loading matrix. indigo, you can replace the unstructured covariance with factor analytic form: * Recall covariances are symmetric so there is no need to estimate the parameters in the upper (or lower) triangle of covariance matrices.įor some. yellow with the number of trials so it quickly becomes too many parameters to estimate. * The number of parameters to be estimated grows. * **G**g is a known genotype relationship matrix. * **G**e may be assumed a general matrix such as unstructured matrix that is usually estimated from the data. Where we assume a separable variance structure for genotype-by-environment effects: indigo (not all genotypes appear in each site). We saw before that we can fit a model that borrows strength across sites for a more *accurate prediction* of genotype by site effects:Īssume that there are. # This work by Emi Tanaka is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.Ĭlass: bg-main1 split-30 hide-slide-number # These slides may take a while to render properly. # Emi Tanaka School of Mathematics and Statisitcs # Statistical Methods for Omics Assisted Breeding Statistical Methods for Omics Assisted BreedingĬlass: split-60 title-slide2 with-border white


 0 kommentar(er)
0 kommentar(er)
